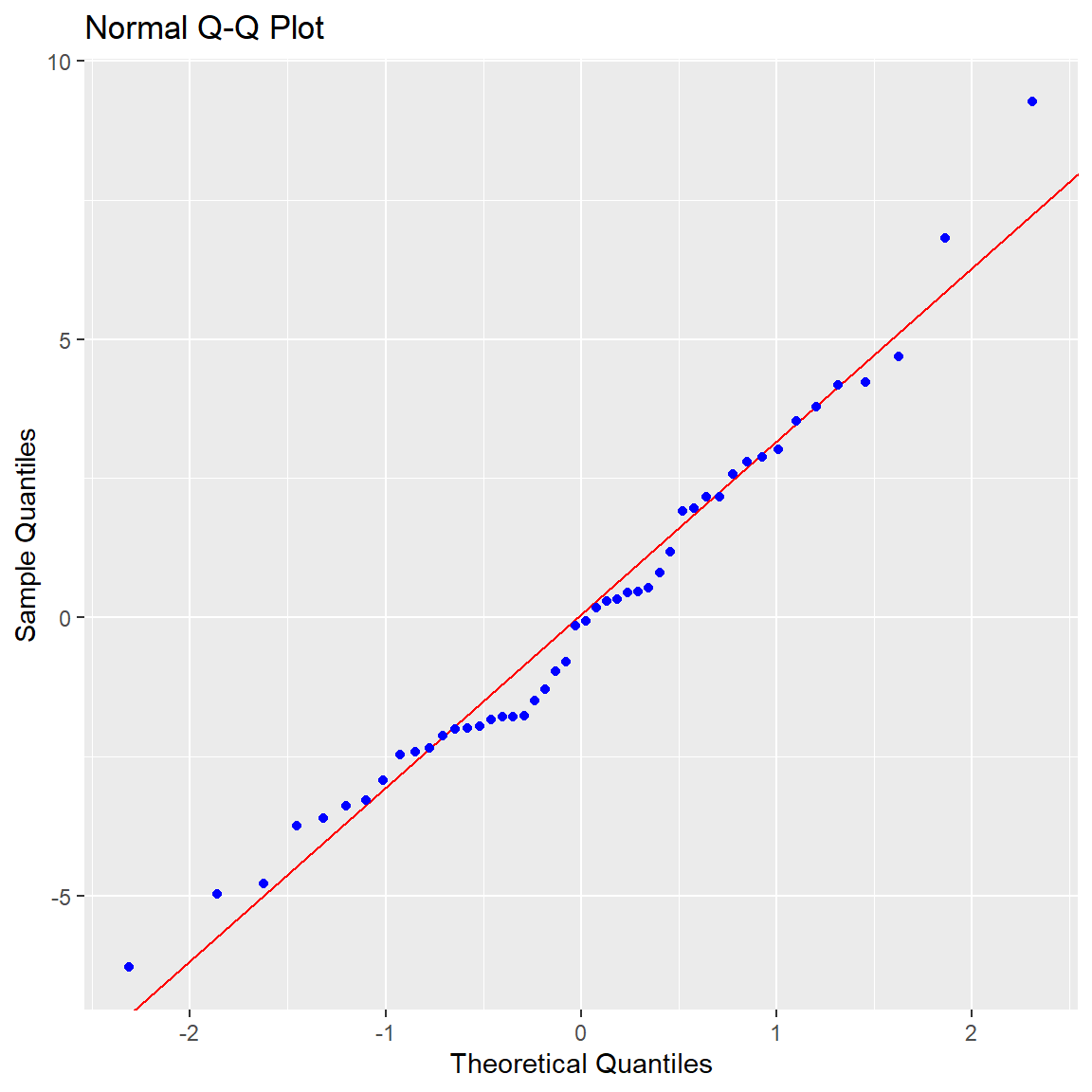
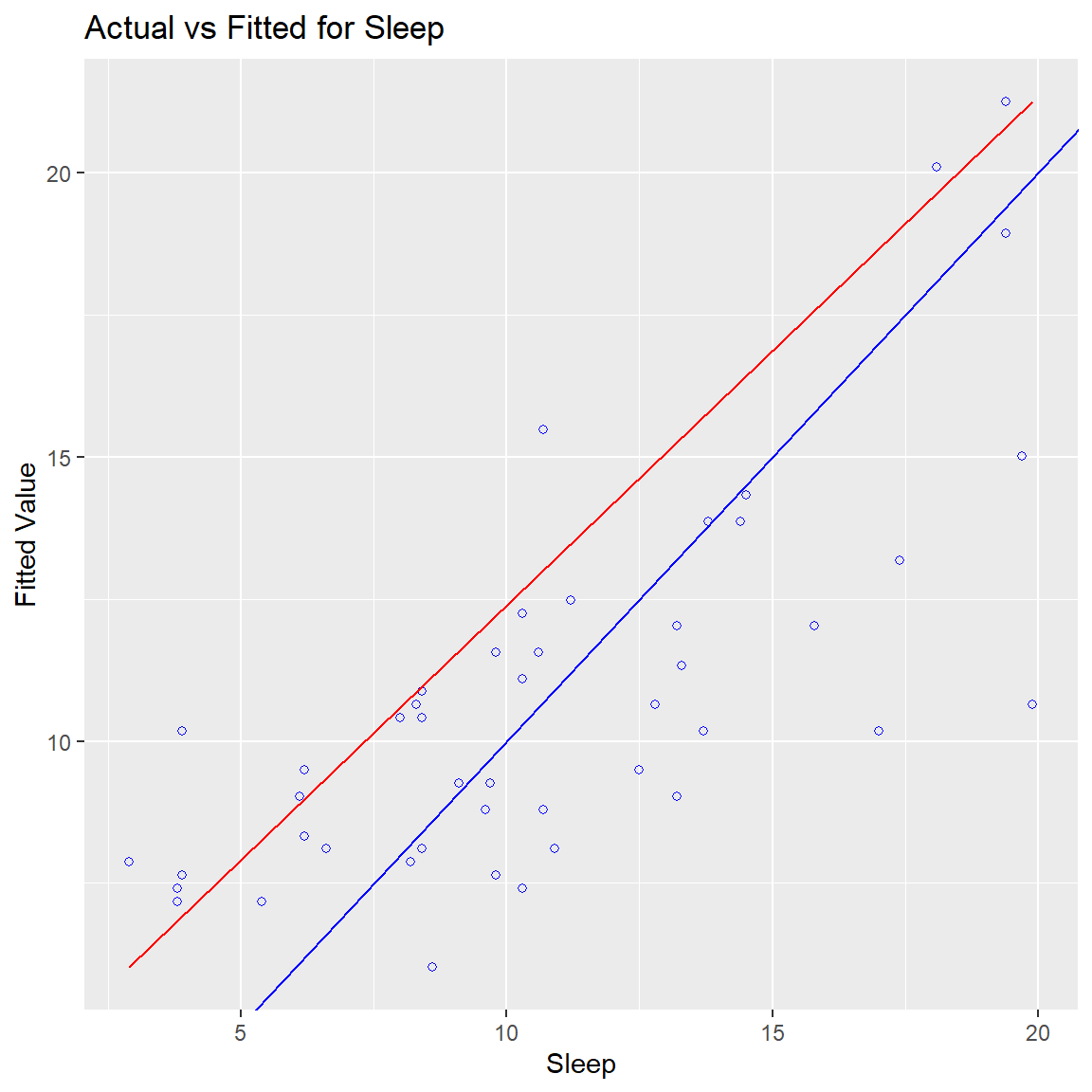
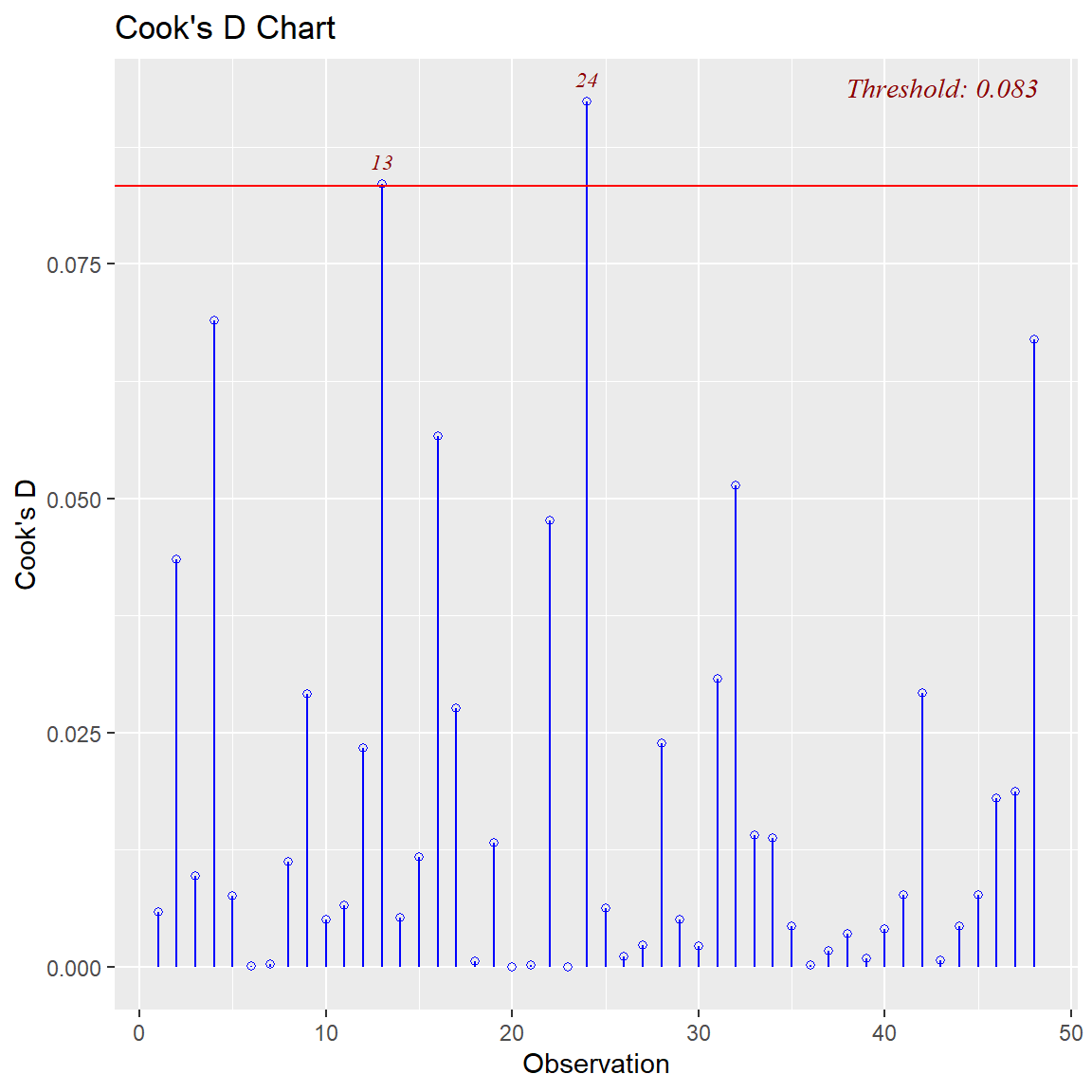
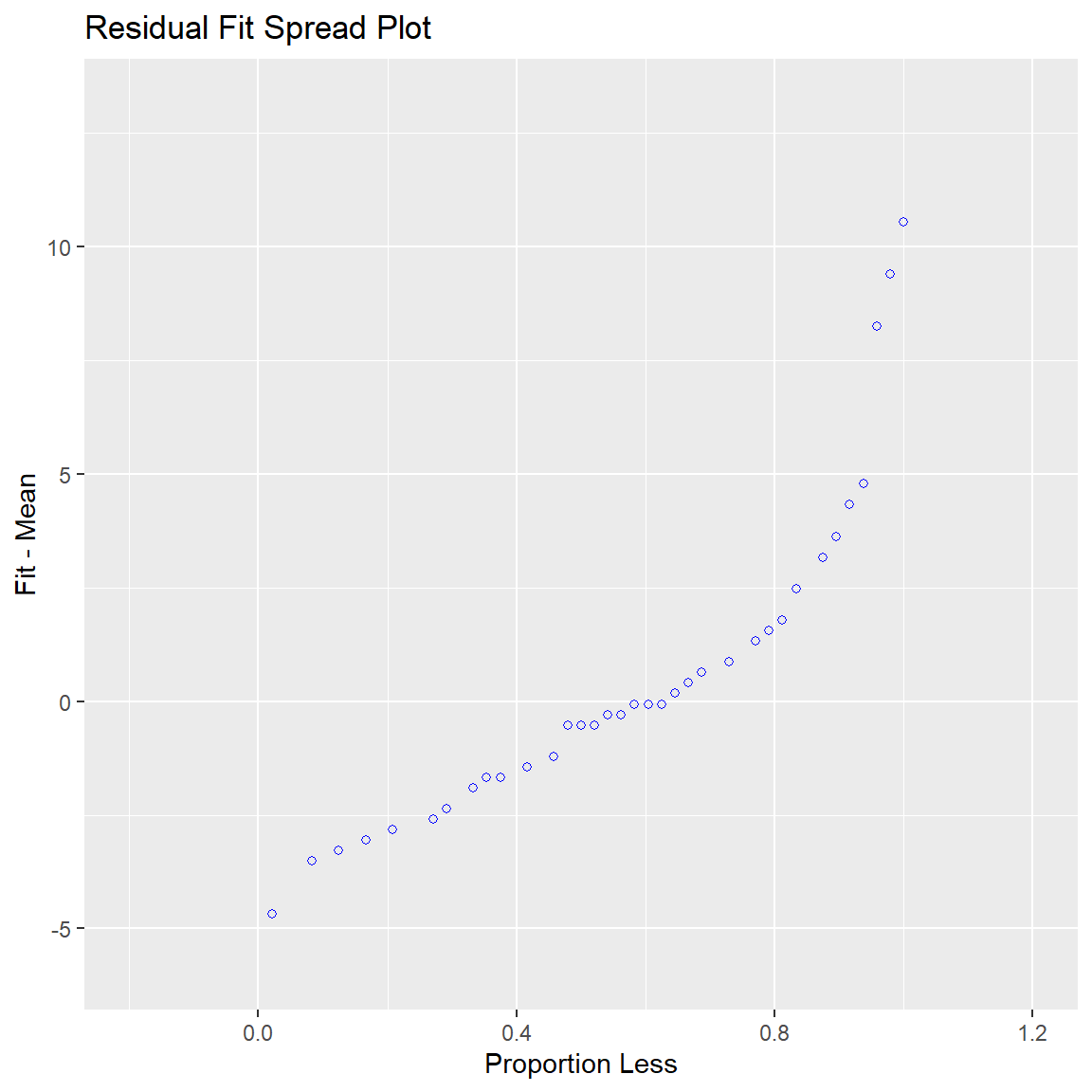
Ben-Shachar, Mattan S., Daniel Lüdecke, and Dominique Makowski. 2020.
“effectsize: Estimation of Effect Size Indices and Standardized Parameters.” Journal of Open Source Software 5 (56): 2815.
https://doi.org/10.21105/joss.02815.
Ben-Shachar, Mattan S., Dominique Makowski, Daniel Lüdecke, Indrajeet Patil, Brenton M. Wiernik, Rémi Thériault, and Philip Waggoner. 2025.
Effectsize: Indices of Effect Size.
https://easystats.github.io/effectsize/.
Fox, John. 2003.
“Effect Displays in R for Generalised Linear Models.” Journal of Statistical Software 8 (15): 1–27.
https://doi.org/10.18637/jss.v008.i15.
Fox, John, and Jangman Hong. 2009.
“Effect Displays in R for Multinomial and Proportional-Odds Logit Models: Extensions to the effects Package.” Journal of Statistical Software 32 (1): 1–24.
https://doi.org/10.18637/jss.v032.i01.
Fox, John, and Sanford Weisberg. 2018.
“Visualizing Fit and Lack of Fit in Complex Regression Models with Predictor Effect Plots and Partial Residuals.” Journal of Statistical Software 87 (9): 1–27.
https://doi.org/10.18637/jss.v087.i09.
———. 2019a.
An R Companion to Applied Regression. Third. Thousand Oaks
CA: Sage.
https://www.john-fox.ca/Companion/.
———. 2019b.
An r Companion to Applied Regression. 3rd ed. Thousand Oaks CA: Sage.
https://socialsciences.mcmaster.ca/jfox/Books/Companion/index.html.
Fox, John, Sanford Weisberg, and Brad Price. 2022.
carData: Companion to Applied Regression Data Sets.
https://r-forge.r-project.org/projects/car/.
———. 2024.
Car: Companion to Applied Regression.
https://r-forge.r-project.org/projects/car/.
Fox, John, Sanford Weisberg, Brad Price, Michael Friendly, and Jangman Hong. 2022.
Effects: Effect Displays for Linear, Generalized Linear, and Other Models.
https://www.r-project.org.
Hebbali, Aravind. 2024.
Olsrr: Tools for Building OLS Regression Models.
https://olsrr.rsquaredacademy.com/.
Iannone, Richard, Joe Cheng, Barret Schloerke, Ellis Hughes, Alexandra Lauer, JooYoung Seo, Ken Brevoort, and Olivier Roy. 2024.
Gt: Easily Create Presentation-Ready Display Tables.
https://gt.rstudio.com.
Ihaka, Ross, Paul Murrell, Kurt Hornik, Jason C. Fisher, Reto Stauffer, Claus O. Wilke, Claire D. McWhite, and Achim Zeileis. 2023.
Colorspace: A Toolbox for Manipulating and Assessing Colors and Palettes.
https://colorspace.R-Forge.R-project.org/.
Kowarik, Alexander, and Matthias Templ. 2016.
“Imputation with the R Package VIM.” Journal of Statistical Software 74 (7): 1–16.
https://doi.org/10.18637/jss.v074.i07.
Lüdecke, Daniel, Mattan S. Ben-Shachar, Indrajeet Patil, and Dominique Makowski. 2020.
“Extracting, Computing and Exploring the Parameters of Statistical Models Using R.” Journal of Open Source Software 5 (53): 2445.
https://doi.org/10.21105/joss.02445.
Lüdecke, Daniel, Mattan S. Ben-Shachar, Indrajeet Patil, Philip Waggoner, and Dominique Makowski. 2021.
“performance: An R Package for Assessment, Comparison and Testing of Statistical Models.” Journal of Open Source Software 6 (60): 3139.
https://doi.org/10.21105/joss.03139.
Lüdecke, Daniel, Mattan S. Ben-Shachar, Indrajeet Patil, Brenton M. Wiernik, Etienne Bacher, Rémi Thériault, and Dominique Makowski. 2022.
“Easystats: Framework for Easy Statistical Modeling, Visualization, and Reporting.” CRAN.
https://doi.org/10.32614/CRAN.package.easystats.
Lüdecke, Daniel, Dominique Makowski, Mattan S. Ben-Shachar, Indrajeet Patil, Søren Højsgaard, and Brenton M. Wiernik. 2025.
Parameters: Processing of Model Parameters.
https://easystats.github.io/parameters/.
Lüdecke, Daniel, Dominique Makowski, Mattan S. Ben-Shachar, Indrajeet Patil, Philip Waggoner, Brenton M. Wiernik, and Rémi Thériault. 2025.
Performance: Assessment of Regression Models Performance.
https://easystats.github.io/performance/.
Lüdecke, Daniel, Dominique Makowski, Mattan S. Ben-Shachar, Indrajeet Patil, Brenton M. Wiernik, Etienne Bacher, and Rémi Thériault. 2025.
Easystats: Framework for Easy Statistical Modeling, Visualization, and Reporting.
https://easystats.github.io/easystats/.
Lüdecke, Daniel, Dominique Makowski, Indrajeet Patil, Mattan S. Ben-Shachar, Brenton M. Wiernik, and Philip Waggoner. 2025.
See: Model Visualisation Toolbox for Easystats and Ggplot2.
https://easystats.github.io/see/.
Lüdecke, Daniel, Dominique Makowski, Indrajeet Patil, Philip Waggoner, Mattan S. Ben-Shachar, Brenton M. Wiernik, Vincent Arel-Bundock, and Etienne Bacher. 2025.
Insight: Easy Access to Model Information for Various Model Objects.
https://easystats.github.io/insight/.
Lüdecke, Daniel, Indrajeet Patil, Mattan S. Ben-Shachar, Brenton M. Wiernik, Philip Waggoner, and Dominique Makowski. 2021.
“see: An R Package for Visualizing Statistical Models.” Journal of Open Source Software 6 (64): 3393.
https://doi.org/10.21105/joss.03393.
Lüdecke, Daniel, Philip Waggoner, and Dominique Makowski. 2019.
“insight: A Unified Interface to Access Information from Model Objects in R.” Journal of Open Source Software 4 (38): 1412.
https://doi.org/10.21105/joss.01412.
Makowski, Dominique, Mattan S. Ben-Shachar, and Daniel Lüdecke. 2019.
“bayestestR: Describing Effects and Their Uncertainty, Existence and Significance Within the Bayesian Framework.” Journal of Open Source Software 4 (40): 1541.
https://doi.org/10.21105/joss.01541.
Makowski, Dominique, Mattan S. Ben-Shachar, Indrajeet Patil, and Daniel Lüdecke. 2020.
“Methods and Algorithms for Correlation Analysis in R.” Journal of Open Source Software 5 (51): 2306.
https://doi.org/10.21105/joss.02306.
Makowski, Dominique, Mattan S. Ben-Shachar, Brenton M. Wiernik, Indrajeet Patil, Rémi Thériault, and Daniel Lüdecke. 2025.
“modelbased: An R Package to Make the Most Out of Your Statistical Models Through Marginal Means, Marginal Effects, and Model Predictions.” Journal of Open Source Software 10 (109): 7969.
https://doi.org/10.21105/joss.07969.
Makowski, Dominique, Daniel Lüdecke, Mattan S. Ben-Shachar, Indrajeet Patil, and Rémi Thériault. 2025.
Modelbased: Estimation of Model-Based Predictions, Contrasts and Means.
https://easystats.github.io/modelbased/.
Makowski, Dominique, Daniel Lüdecke, Mattan S. Ben-Shachar, Indrajeet Patil, Micah K. Wilson, and Brenton M. Wiernik. 2025.
bayestestR: Understand and Describe Bayesian Models and Posterior Distributions.
https://easystats.github.io/bayestestR/.
Makowski, Dominique, Daniel Lüdecke, Indrajeet Patil, Rémi Thériault, Mattan S. Ben-Shachar, and Brenton M. Wiernik. 2023.
“Automated Results Reporting as a Practical Tool to Improve Reproducibility and Methodological Best Practices Adoption.” CRAN.
https://easystats.github.io/report/.
———. 2025.
Report: Automated Reporting of Results and Statistical Models.
https://easystats.github.io/report/.
Makowski, Dominique, Brenton M. Wiernik, Indrajeet Patil, Daniel Lüdecke, and Mattan S. Ben-Shachar. 2022.
“correlation: Methods for Correlation Analysis.” https://CRAN.R-project.org/package=correlation.
Makowski, Dominique, Brenton M. Wiernik, Indrajeet Patil, Daniel Lüdecke, Mattan S. Ben-Shachar, and Rémi Thériault. 2025.
Correlation: Methods for Correlation Analysis.
https://easystats.github.io/correlation/.
Patil, Indrajeet, Etienne Bacher, Dominique Makowski, Daniel Lüdecke, Mattan S. Ben-Shachar, and Brenton M. Wiernik. 2025.
Datawizard: Easy Data Wrangling and Statistical Transformations.
https://easystats.github.io/datawizard/.
Patil, Indrajeet, Dominique Makowski, Mattan S. Ben-Shachar, Brenton M. Wiernik, Etienne Bacher, and Daniel Lüdecke. 2022.
“datawizard: An R Package for Easy Data Preparation and Statistical Transformations.” Journal of Open Source Software 7 (78): 4684.
https://doi.org/10.21105/joss.04684.
R Core Team. 2025.
R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing.
https://www.R-project.org/.
Robinson, David, Alex Hayes, and Simon Couch. 2025.
Broom: Convert Statistical Objects into Tidy Tibbles.
https://broom.tidymodels.org/.
Sing, Tobias, Oliver Sander, Niko Beerenwinkel, and Thomas Lengauer. 2020.
ROCR: Visualizing the Performance of Scoring Classifiers.
http://ipa-tys.github.io/ROCR/.
Sing, T., O. Sander, N. Beerenwinkel, and T. Lengauer. 2005.
“ROCR: Visualizing Classifier Performance in r.” Bioinformatics 21 (20): 7881.
http://rocr.bioinf.mpi-sb.mpg.de.
Sjoberg, Daniel D., Joseph Larmarange, Michael Curry, Jessica Lavery, Karissa Whiting, and Emily C. Zabor. 2024.
Gtsummary: Presentation-Ready Data Summary and Analytic Result Tables.
https://github.com/ddsjoberg/gtsummary.
Sjoberg, Daniel D., Karissa Whiting, Michael Curry, Jessica A. Lavery, and Joseph Larmarange. 2021.
“Reproducible Summary Tables with the Gtsummary Package.” The R Journal 13: 570–80.
https://doi.org/10.32614/RJ-2021-053.
Stauffer, Reto, Georg J. Mayr, Markus Dabernig, and Achim Zeileis. 2009.
“Somewhere over the Rainbow: How to Make Effective Use of Colors in Meteorological Visualizations.” Bulletin of the American Meteorological Society 96 (2): 203–16.
https://doi.org/10.1175/BAMS-D-13-00155.1.
Templ, Matthias, Alexander Kowarik, Andreas Alfons, Gregor de Cillia, and Wolfgang Rannetbauer. 2022.
VIM: Visualization and Imputation of Missing Values.
https://github.com/statistikat/VIM.
Wickham, Hadley, Romain François, Lionel Henry, Kirill Müller, and Davis Vaughan. 2023.
Dplyr: A Grammar of Data Manipulation.
https://dplyr.tidyverse.org.
Zeileis, Achim, Jason C. Fisher, Kurt Hornik, Ross Ihaka, Claire D. McWhite, Paul Murrell, Reto Stauffer, and Claus O. Wilke. 2020.
“colorspace: A Toolbox for Manipulating and Assessing Colors and Palettes.” Journal of Statistical Software 96 (1): 1–49.
https://doi.org/10.18637/jss.v096.i01.
Zeileis, Achim, Kurt Hornik, and Paul Murrell. 2009.
“Escaping RGBland: Selecting Colors for Statistical Graphics.” Computational Statistics & Data Analysis 53 (9): 3259–70.
https://doi.org/10.1016/j.csda.2008.11.033.